T-test vs Wilcoxon vs Median Test
Define a list of laws (i.e., distributions) with mean and sd.
# functions
n <- 40 # per group
laws <- list(
norm = function(mean=0, sd=1) mean + sd * rnorm(n, 0, 1),
logn = function(mean=0, sd=1) mean + sd * (rlnorm(n, sdlog=1) - 1.65) / 1.95,
exp1 = function(mean=0, sd=1) mean + sd * (rexp(n) - 1),
exp2 = function(mean=0, sd=1) mean + sd * (rexp(n)^2 - 2) / 3.94,
emix = function(mean=0, sd=1) mean + sd * (c(rep(0, n/2), rexp(n/2)) - 0.5) / 0.84
)
set.seed(123)
options(digits=3)
get_data_all_laws <- function(){
dat <- lapply(names(laws), function(fname){
f <- laws[[fname]]
data.frame(y=f(), fun=fname)
})
dat <- do.call(rbind, dat)
dat
}
boxplot(y ~ fun, get_data_all_laws(), main= "Laws illustrated")
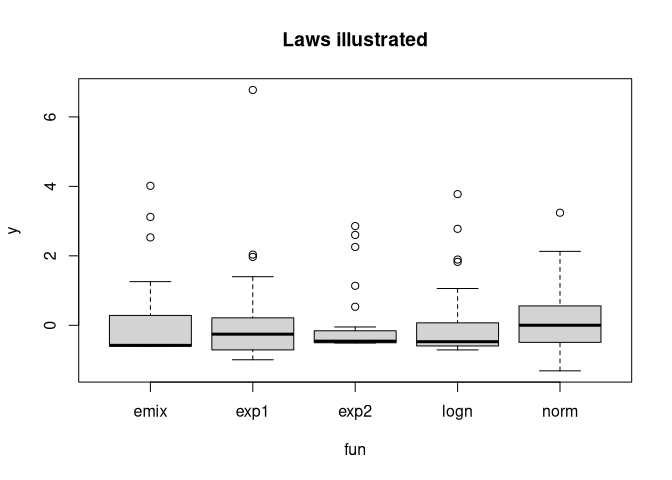
Verify mean and standard deviation
sapply(laws, function(f) mean(replicate(10000, mean(f(0, 1)))))
norm logn exp1 exp2 emix
1.55e-03 -2.89e-03 -1.66e-03 -8.53e-05 -1.32e-03
sapply(laws, function(f) mean(replicate(10000, mean(f(1, 2)))))
norm logn exp1 exp2 emix
1.003 1.002 0.998 0.994 0.997
sapply(laws, function(f) mean(replicate(10000, sd(f(0, 1)))))
norm logn exp1 exp2 emix
0.994 1.008 0.977 0.999 1.000
sapply(laws, function(f) mean(replicate(10000, sd(f(1, 2)))))
norm logn exp1 exp2 emix
1.99 2.01 1.96 1.99 2.00
Define the tests used
pval_t <- function(d) t.test(y ~ group,d)$p.value
pval_w <- function(d) coin::pvalue(coin::wilcox_test(y ~ group, d))
pval_m <- function(d) coin::pvalue(coin::median_test(y ~ group, d))
# retuns data with f1() for group "A" and f2(mean2, sd2) for group "B"
get_data <- function(f1, f2, mean2=0, sd2=1){
d <- rbind(
data.frame(
y=f1(),
group="A"
),
data.frame(
y=f2(mean2, sd2),
group="B"
))
d$group <- as.factor(d$group)
d
}
# get power of tests
get_power <- function(f1, f2, nsim=1000, mean2=0, sd2=1) {
data_list <- replicate(nsim, get_data(f1, f2, mean2=mean2, sd2=sd2), simplify = FALSE)
c(t = mean(sapply(data_list, pval_t) < 0.05),
w = mean(sapply(data_list, pval_w) < 0.05),
m = mean(sapply(data_list, pval_m) < 0.05))
}
# get all combinations of functions
fun_comb <- expand.grid(names(laws), names(laws)) |> as.matrix()
rnames <- apply(fun_comb, 1, paste0, collapse="_")
sim <- function(nsim=1000, mean2=0, sd2=1) {
fun_comb_list <- split(fun_comb, row(fun_comb))
coverage <- parallel::mclapply(fun_comb_list, function(f_names){
f1 <- laws[[f_names[1]]]
f2 <- laws[[f_names[2]]]
get_power(f1, f2, nsim=nsim, mean2=mean2, sd2=sd2)
})
coverage <- do.call(rbind, coverage)
rownames(coverage) <- rnames
colnames(coverage) <- paste0(
colnames(coverage), " ", as.character(mean2), " ", as.character(sd2))
coverage |> as.data.frame()
}
Simulation
nsim <- 500
set.seed(4321)
null <- sim(nsim=nsim)
set.seed(4321)
s_diff <- sim(nsim=nsim, sd2=2)
set.seed(4321)
s_difff <- sim(nsim=nsim, sd2=5)
set.seed(4321)
mu_diff <- sim(nsim=nsim, mean2 = 0.1)
set.seed(4321)
mu_difff <- sim(nsim=nsim, mean2 = 0.2)
results <- cbind(
null,
s_diff,
s_difff,
mu_diff,
mu_difff
)
results * 100
| t 0 1 | w 0 1 | m 0 1 | t 0 2 | w 0 2 | m 0 2 | t 0 5 | w 0 5 | m 0 5 | t 0.1 1 | w 0.1 1 | m 0.1 1 | t 0.2 1 | w 0.2 1 | m 0.2 1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| norm_norm | 4.2 | 4.4 | 3.6 | 4.4 | 4.6 | 5.0 | 7.0 | 10.6 | 10.6 | 8.4 | 7.6 | 4.6 | 14.4 | 12.4 | 8.4 |
| logn_norm | 8.0 | 12.6 | 33.6 | 4.2 | 8.0 | 16.4 | 4.8 | 9.0 | 14.6 | 12.8 | 25.4 | 45.6 | 21.0 | 41.4 | 59.8 |
| exp1_norm | 5.8 | 9.8 | 21.4 | 3.8 | 5.6 | 13.6 | 2.6 | 6.6 | 11.6 | 8.6 | 17.4 | 29.4 | 17.8 | 32.2 | 45.8 |
| exp2_norm | 5.6 | 15.4 | 46.8 | 5.6 | 11.4 | 26.0 | 3.4 | 10.6 | 16.2 | 11.4 | 27.2 | 63.2 | 21.6 | 46.4 | 79.6 |
| emix_norm | 3.8 | 10.2 | 46.4 | 5.6 | 9.6 | 25.4 | 4.8 | 7.8 | 11.2 | 10.0 | 23.8 | 61.0 | 15.6 | 42.4 | 80.4 |
| norm_logn | 5.2 | 14.2 | 31.0 | 7.8 | 41.0 | 54.0 | 7.4 | 77.0 | 76.4 | 4.0 | 4.8 | 15.8 | 12.0 | 7.8 | 9.0 |
| logn_logn | 5.4 | 7.8 | 5.8 | 7.6 | 56.8 | 32.6 | 10.6 | 77.6 | 70.2 | 6.8 | 17.4 | 8.4 | 14.0 | 51.4 | 25.8 |
| exp1_logn | 2.8 | 6.0 | 4.8 | 8.4 | 50.4 | 28.0 | 10.8 | 77.2 | 64.8 | 6.2 | 20.2 | 7.8 | 14.4 | 41.6 | 18.6 |
| exp2_logn | 4.4 | 7.6 | 7.8 | 5.8 | 58.8 | 33.8 | 11.8 | 80.8 | 73.2 | 8.8 | 9.8 | 23.4 | 17.4 | 52.0 | 54.0 |
| emix_logn | 2.6 | 4.4 | 10.0 | 5.4 | 55.0 | 0.8 | 11.8 | 76.0 | 17.0 | 3.8 | 35.0 | 31.0 | 11.6 | 92.8 | 56.6 |
| norm_exp1 | 3.8 | 9.4 | 17.6 | 6.0 | 29.6 | 34.4 | 8.2 | 49.6 | 51.2 | 5.6 | 6.2 | 11.6 | 10.8 | 7.2 | 5.8 |
| logn_exp1 | 5.0 | 7.8 | 5.0 | 4.8 | 33.0 | 17.8 | 6.8 | 51.0 | 44.2 | 10.8 | 6.6 | 9.0 | 17.8 | 23.2 | 18.6 |
| exp1_exp1 | 4.4 | 4.6 | 4.8 | 5.6 | 30.0 | 14.8 | 7.0 | 48.4 | 38.6 | 7.6 | 10.2 | 8.0 | 12.6 | 28.2 | 13.0 |
| exp2_exp1 | 5.2 | 11.2 | 9.2 | 4.6 | 33.8 | 18.6 | 6.6 | 49.0 | 44.4 | 9.8 | 6.0 | 20.2 | 20.8 | 21.0 | 44.6 |
| emix_exp1 | 2.8 | 3.8 | 10.8 | 4.8 | 34.2 | 1.0 | 6.6 | 50.4 | 5.4 | 4.6 | 7.0 | 20.0 | 12.2 | 29.2 | 46.2 |
| norm_exp2 | 5.2 | 15.4 | 48.0 | 7.8 | 51.0 | 80.6 | 14.6 | 95.0 | 94.4 | 3.6 | 8.4 | 29.4 | 9.2 | 7.2 | 17.2 |
| logn_exp2 | 2.8 | 9.0 | 6.8 | 7.6 | 86.2 | 67.8 | 14.8 | 97.2 | 94.6 | 6.2 | 38.2 | 10.6 | 17.6 | 69.4 | 27.4 |
| exp1_exp2 | 4.4 | 10.4 | 7.6 | 8.6 | 70.4 | 57.2 | 14.2 | 97.4 | 93.0 | 6.2 | 24.6 | 7.2 | 12.8 | 47.2 | 16.2 |
| exp2_exp2 | 4.2 | 5.6 | 5.0 | 9.4 | 91.0 | 70.8 | 11.0 | 94.8 | 92.2 | 5.8 | 67.8 | 23.2 | 12.4 | 91.0 | 59.6 |
| emix_exp2 | 2.0 | 64.8 | 9.0 | 9.0 | 87.0 | 7.4 | 14.4 | 95.4 | 48.6 | 3.6 | 85.2 | 35.8 | 9.8 | 96.6 | 61.0 |
| norm_emix | 3.2 | 10.6 | 47.2 | 2.4 | 49.6 | 77.0 | 4.0 | 91.4 | 90.4 | 3.6 | 6.6 | 32.0 | 9.6 | 4.0 | 18.2 |
| logn_emix | 1.8 | 2.4 | 9.4 | 3.6 | 87.0 | 61.8 | 4.0 | 90.4 | 86.0 | 8.2 | 9.0 | 2.2 | 15.4 | 43.6 | 0.4 |
| exp1_emix | 3.0 | 5.0 | 10.8 | 2.2 | 85.8 | 53.4 | 4.4 | 92.2 | 87.4 | 3.2 | 14.2 | 4.2 | 11.4 | 35.0 | 1.0 |
| exp2_emix | 1.2 | 62.0 | 9.8 | 2.4 | 87.0 | 65.8 | 4.2 | 91.2 | 88.8 | 5.6 | 1.0 | 0.8 | 18.4 | 71.6 | 0.0 |
| emix_emix | 1.2 | 0.0 | 0.0 | 2.8 | 89.2 | 2.0 | 3.8 | 91.6 | 20.2 | 2.4 | 82.4 | 0.0 | 10.0 | 94.8 | 0.2 |
Confidence intervals of ratios
prop <- function(ratio, nsim){
confint_ <- prop.test(round(ratio*nsim), nsim)$conf.int[1:2]
names(confint_) <- c("lower", "upper")
c(
ratio= ratio,
confint_
)}
sapply(c(0:4/40, 3:10/20), prop, nsim) |> as.data.frame() * 100
| V1 | V2 | V3 | V4 | V5 | V6 | V7 | V8 | V9 | V10 | V11 | V12 | V13 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ratio | 0.00 | 2.50 | 5.00 | 7.5 | 10.00 | 15.0 | 20.0 | 25.0 | 30.0 | 35.0 | 40.0 | 45.0 | 50.0 |
| lower | 0.00 | 1.30 | 3.33 | 5.5 | 7.58 | 12.0 | 16.6 | 21.3 | 26.1 | 30.9 | 35.7 | 40.6 | 45.6 |
| upper | 0.95 | 4.27 | 7.39 | 10.4 | 13.05 | 18.5 | 23.8 | 29.1 | 34.3 | 39.4 | 44.5 | 49.5 | 54.4 |
this gives an idea of the uncertainty of a ratio given 500 simulations
redo analysis but with groupwise equal medians
Define a list of laws (i.e., distributions) with median and sd
# functions
n <- 40 # per group
# mean == to keep notation consistent
laws <- list(
norm = function(mean=0, sd=1) mean + sd * rnorm(n, 0, 1)
,logn = function(mean=0, sd=1) mean + sd * (rlnorm(n, sdlog=1) - 1.02) / 1.95
,exp1 = function(mean=0, sd=1) mean + sd * (rexp(n) - 0.706)
,exp2 = function(mean=0, sd=1) mean + sd * (rexp(n)^2 - 0.523) / 3.94
,emix = function(mean=0, sd=1) mean + sd * (c(rep(0, n/2), rexp(n/2)) -0.025) / 0.84
)
verify median and standard deviation
sapply(laws, function(f) mean(replicate(10000, median(f(0, 1)))))
norm logn exp1 exp2 emix
-0.000142 -0.000216 0.002144 0.000204 0.000246
sapply(laws, function(f) mean(replicate(10000, median(f(1, 2)))))
norm logn exp1 exp2 emix
1.000 1.001 0.998 1.000 0.999
sapply(laws, function(f) mean(replicate(10000, sd(f(0, 1)))))
norm logn exp1 exp2 emix
0.996 1.006 0.978 1.010 1.003
sapply(laws, function(f) mean(replicate(10000, sd(f(1, 2)))))
norm logn exp1 exp2 emix
1.99 2.00 1.95 1.99 2.01
boxplot(y ~ fun, get_data_all_laws(), main= "Equal Medians (in expectation)")
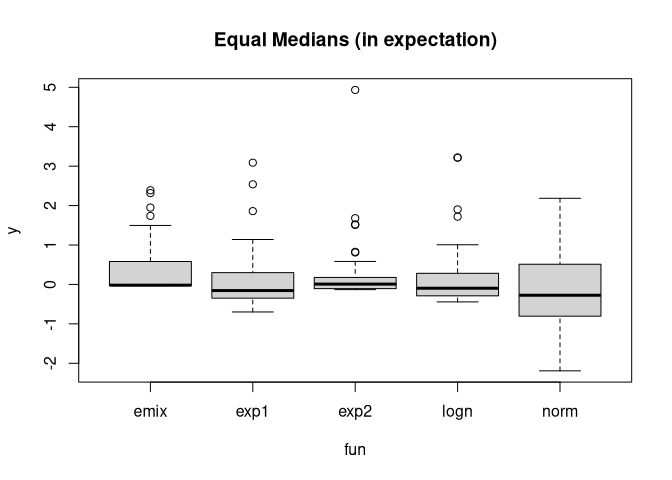
set.seed(4321)
null_median <- sim(nsim=nsim)
set.seed(4321)
s_diff_median <- sim(nsim=nsim, sd2=2)
set.seed(4321)
s_difff_median <- sim(nsim=nsim, sd2=5)
set.seed(4321)
mu_diff_median <- sim(nsim=nsim, mean2 = 0.1)
set.seed(4321)
mu_difff_median <- sim(nsim=nsim, mean2 = 0.2)
results_median <- cbind(
null_median,
s_diff_median,
s_difff_median,
mu_diff_median,
mu_difff_median
)
results_median * 100
| t 0 1 | w 0 1 | m 0 1 | t 0 2 | w 0 2 | m 0 2 | t 0 5 | w 0 5 | m 0 5 | t 0.1 1 | w 0.1 1 | m 0.1 1 | t 0.2 1 | w 0.2 1 | m 0.2 1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| norm_norm | 4.4 | 4.0 | 3.6 | 4.8 | 5.6 | 6.0 | 3.8 | 8.4 | 9.4 | 7.8 | 6.6 | 7.0 | 14.8 | 15.2 | 10.2 |
| logn_norm | 22.0 | 13.0 | 4.6 | 13.4 | 12.6 | 7.0 | 7.6 | 9.8 | 10.2 | 13.2 | 8.0 | 7.2 | 6.2 | 5.8 | 16.6 |
| exp1_norm | 20.8 | 10.6 | 3.4 | 12.6 | 11.6 | 6.0 | 5.4 | 8.2 | 9.4 | 11.8 | 6.4 | 6.0 | 7.8 | 7.2 | 12.2 |
| exp2_norm | 36.0 | 23.6 | 7.6 | 16.6 | 16.0 | 12.0 | 7.8 | 15.2 | 13.0 | 19.0 | 11.4 | 8.8 | 7.2 | 5.6 | 18.6 |
| emix_norm | 79.6 | 58.2 | 0.8 | 31.6 | 29.2 | 4.2 | 13.0 | 18.4 | 6.4 | 50.2 | 29.8 | 1.4 | 32.2 | 17.4 | 6.2 |
| norm_logn | 19.0 | 11.2 | 6.6 | 36.0 | 12.8 | 5.2 | 47.8 | 9.4 | 6.8 | 45.2 | 32.2 | 7.0 | 61.4 | 48.8 | 14.6 |
| logn_logn | 3.8 | 5.8 | 5.4 | 6.6 | 7.8 | 4.0 | 24.4 | 11.4 | 8.6 | 6.0 | 15.8 | 8.8 | 18.6 | 54.4 | 29.4 |
| exp1_logn | 3.4 | 8.6 | 5.2 | 9.2 | 5.6 | 5.0 | 23.8 | 9.2 | 6.8 | 6.8 | 23.6 | 7.6 | 13.8 | 47.4 | 18.8 |
| exp2_logn | 4.6 | 21.6 | 6.4 | 6.2 | 19.4 | 8.6 | 22.4 | 15.6 | 12.4 | 5.2 | 5.8 | 14.6 | 11.8 | 26.4 | 38.6 |
| emix_logn | 22.2 | 65.4 | 0.6 | 5.2 | 39.8 | 1.2 | 12.8 | 18.8 | 3.0 | 8.6 | 22.2 | 0.4 | 3.8 | 3.4 | 8.2 |
| norm_exp1 | 24.8 | 13.0 | 4.2 | 32.6 | 9.2 | 5.4 | 39.8 | 7.4 | 9.0 | 44.2 | 25.8 | 5.8 | 57.2 | 43.6 | 8.8 |
| logn_exp1 | 3.0 | 7.4 | 5.4 | 8.4 | 12.8 | 8.0 | 23.4 | 12.6 | 12.2 | 8.8 | 7.6 | 9.6 | 16.8 | 18.4 | 18.0 |
| exp1_exp1 | 5.2 | 5.8 | 5.8 | 9.0 | 8.8 | 6.8 | 24.4 | 9.6 | 10.8 | 8.4 | 10.2 | 6.8 | 13.6 | 26.4 | 11.8 |
| exp2_exp1 | 3.8 | 17.6 | 6.6 | 6.8 | 20.2 | 10.6 | 23.4 | 10.0 | 11.8 | 4.4 | 8.4 | 9.6 | 9.2 | 5.2 | 19.2 |
| emix_exp1 | 18.6 | 53.4 | 0.6 | 5.0 | 32.2 | 2.2 | 15.2 | 15.4 | 5.0 | 9.4 | 28.8 | 2.2 | 4.0 | 11.4 | 5.2 |
| norm_exp2 | 34.0 | 23.6 | 7.4 | 59.4 | 35.4 | 3.6 | 73.6 | 30.0 | 6.6 | 54.6 | 43.8 | 11.8 | 72.2 | 58.0 | 21.4 |
| logn_exp2 | 3.4 | 25.4 | 6.2 | 15.4 | 13.2 | 3.6 | 45.2 | 8.8 | 8.6 | 9.8 | 55.6 | 18.2 | 23.0 | 81.4 | 47.4 |
| exp1_exp2 | 6.0 | 21.8 | 6.8 | 13.8 | 25.0 | 4.0 | 45.2 | 5.8 | 4.4 | 8.6 | 47.4 | 15.6 | 23.2 | 66.6 | 29.2 |
| exp2_exp2 | 3.0 | 5.2 | 4.4 | 8.6 | 17.4 | 6.8 | 40.2 | 14.4 | 10.8 | 5.8 | 62.0 | 22.2 | 15.0 | 92.4 | 66.4 |
| emix_exp2 | 17.4 | 61.0 | 0.0 | 3.8 | 51.6 | 0.4 | 21.2 | 29.2 | 1.4 | 5.4 | 6.8 | 1.8 | 1.6 | 64.2 | 11.4 |
| norm_emix | 75.0 | 56.6 | 0.8 | 97.6 | 78.6 | 0.2 | 100.0 | 89.6 | 0.0 | 89.2 | 74.2 | 1.0 | 97.0 | 88.8 | 2.6 |
| logn_emix | 23.8 | 62.8 | 0.2 | 65.8 | 74.6 | 0.0 | 98.8 | 70.4 | 0.0 | 32.6 | 83.8 | 3.8 | 58.0 | 97.0 | 8.4 |
| exp1_emix | 24.4 | 57.0 | 0.2 | 72.2 | 72.4 | 0.2 | 99.4 | 83.2 | 0.0 | 34.0 | 72.4 | 1.6 | 55.8 | 87.8 | 4.6 |
| exp2_emix | 17.0 | 64.4 | 0.2 | 58.4 | 66.2 | 0.0 | 96.6 | 0.0 | 0.0 | 31.4 | 93.6 | 6.2 | 47.0 | 99.0 | 21.4 |
| emix_emix | 1.4 | 0.0 | 0.0 | 25.6 | 6.0 | 0.0 | 92.2 | 0.2 | 0.0 | 3.0 | 85.2 | 0.0 | 11.4 | 94.4 | 0.4 |